Single-cell Metabolic Profiling
The cellular metabolic processes carry rich and unique information, and are challenging to analyze at the single-cell level. We seek to resolve the metabolic heterogeneity from complex biological samples, and strive to understand the interplay between metabolic and protein signaling activities.
Surface-immobilized fluorescent sensors
We design special fluorescent probes that are specific for target metabolites.
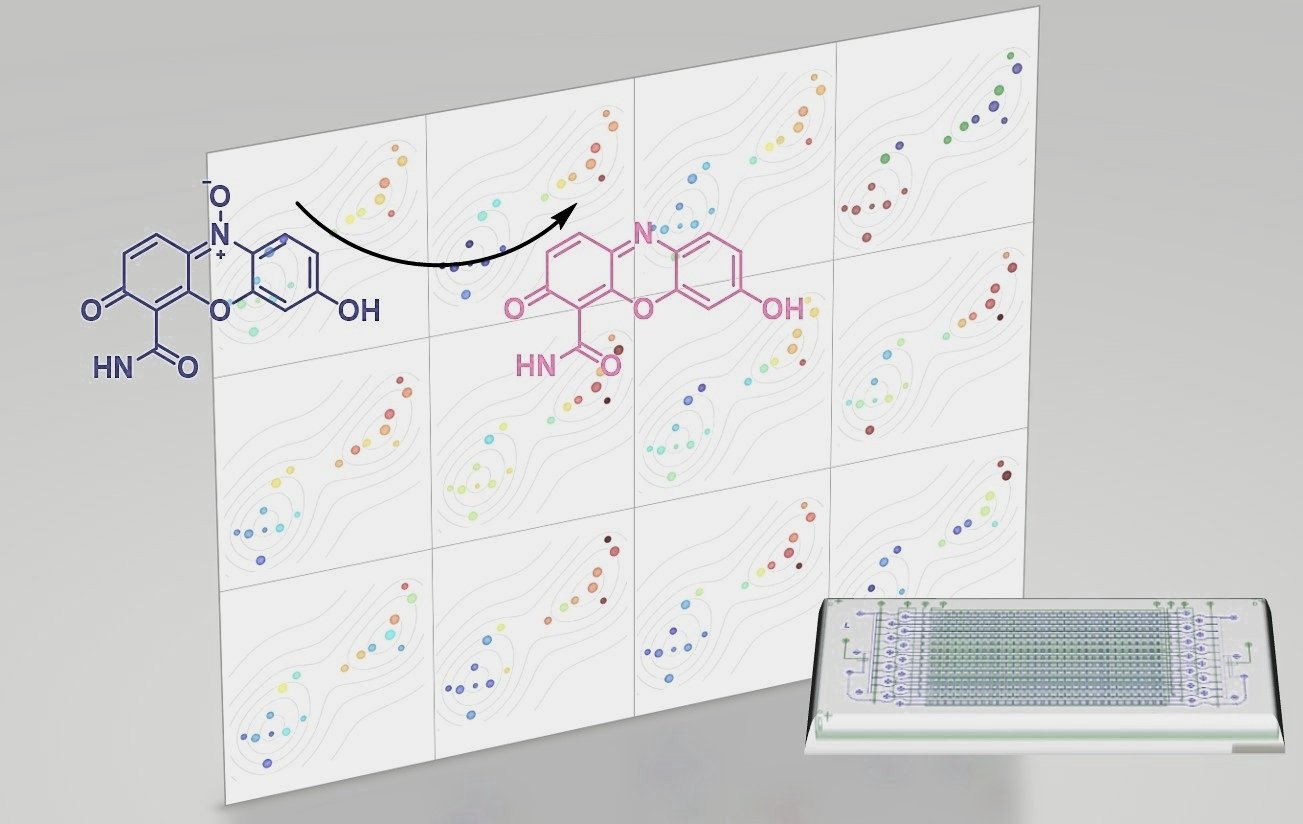
In the example below, we grafted resazurin analogs to the surface and employed coupled enzyme reactions to quantify the lactate production from single cancer cells.
Immuno-mimetic Competitive binding assays
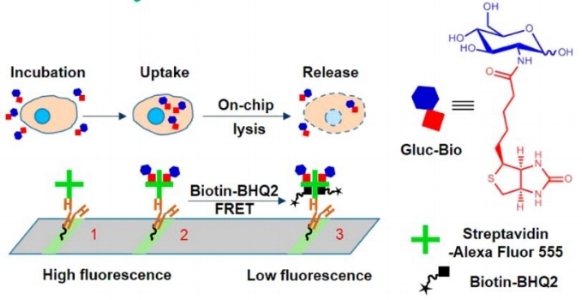
Another approach to quantify metabolites is through surface-based competitive binding assays. The general scheme is rather simple, as shown below.
We have demonstrated a few examples, using this competitive binding principle.
The assay for glutathione (GSH), cAMP and cGMP.
The assay for glucose uptake.
The assay for glutamine uptake.
Single-cell chips
We rely on microfluidic chips to achieve single-cell resolution. These chips have large numbers of micro-chambers that separate individual cells. Cells can be cultured and lysed on the chips. Reagents can be conveniently introduced through microfluidc channels. The implementation of the metabolite assays (described above) on this platform enables the multiplex analytes quantitation from single cells.